Rapid diagnostics of novel coronavirus infection by loop-mediated isothermal amplification
- Authors: Khafizov K.F.1, Petrov V.V.1, Krasovitov K.V.1, Zolkina M.V.1, Akimkin V.G.1
-
Affiliations:
- FSBI Central Research Institute for Epidemiology of the Federal Service for Surveillance of Consumer Rights Protection and Human Wellbeing (Rospotrebnadzor)
- Issue: Vol 66, No 1 (2021)
- Pages: 17-28
- Section: REVIEWS
- URL: https://journal-vniispk.ru/0507-4088/article/view/118153
- DOI: https://doi.org/10.36233/0507-4088-42
- ID: 118153
Cite item
Abstract
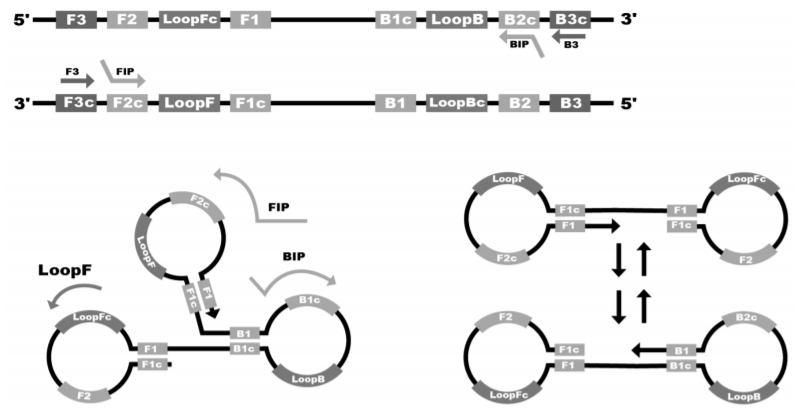
This review presents the basic principles of application of the loop-mediated isothermal amplification (LAMP) reaction for the rapid diagnosis of coronavirus infection caused by SARS-CoV-2. The basic technical details of the method, and the most popular approaches of specific and non-specific detection of amplification products are briefly described. We also discuss the first published works on the use of the method for the detection of the nucleic acid of the SARS-CoV-2 virus, including those being developed in the Russian Federation. For commercially available and published LAMP-based assays, the main analytical characteristics of the tests are listed, which are often comparable to those based on the method of reverse transcription polymerase chain reaction (RT-PCR), and in some cases are even superior. The advantages and limitations of this promising methodology in comparison to other methods of molecular diagnostics, primarily RT-PCR, are discussed, as well as the prospects for the development of technology for the detection of other infectious agents.
Full Text
##article.viewOnOriginalSite##About the authors
K. F. Khafizov
FSBI Central Research Institute for Epidemiology of the Federal Service for Surveillance of Consumer Rights Protection and Human Wellbeing (Rospotrebnadzor)
Author for correspondence.
Email: kkhafizov@gmail.com
ORCID iD: 0000-0001-5524-0296
Khafizov Kamil Faridovich - Ph.D. (Biol.), Head of the Scientific Group for the Development of New Diagnostic Methods of the Department for Molecular Diagnostics and Epidemiology.
111123, Moscow, Novogireevskaya str. 3a
Russian FederationV. V. Petrov
FSBI Central Research Institute for Epidemiology of the Federal Service for Surveillance of Consumer Rights Protection and Human Wellbeing (Rospotrebnadzor)
Email: petrov@pcr.ms
ORCID iD: 0000-0002-3503-2366
Petrov Vadim Viktorovich - Head of the research group for the development of new molecular biological technologies.
111123, Moscow, Novogireevskaya str. 3a
Russian FederationK. V. Krasovitov
FSBI Central Research Institute for Epidemiology of the Federal Service for Surveillance of Consumer Rights Protection and Human Wellbeing (Rospotrebnadzor)
Email: krasovitov@cmd.su
ORCID iD: 0000-0001-7237-1810
Krasovitov Kirill Vladimirovich - Laboratory research assistant, Research group for the development of new molecular biological technologies.
111123, Moscow, Novogireevskaya str. 3a
Russian FederationM. V. Zolkina
FSBI Central Research Institute for Epidemiology of the Federal Service for Surveillance of Consumer Rights Protection and Human Wellbeing (Rospotrebnadzor)
Email: mvzolkina@mail.ru
ORCID iD: 0000-0002-2973-9947
Zolkina Maria Vladimirovna - Head of the development.
111123 Moscow, Novogireevskaya str. 3a
Russian FederationV. G. Akimkin
FSBI Central Research Institute for Epidemiology of the Federal Service for Surveillance of Consumer Rights Protection and Human Wellbeing (Rospotrebnadzor)
Email: vgakimkin@yandex.ru
ORCID iD: 0000-0003-4228-9044
Akimkin Vasily Gennadevich - Academician of the Russian Academy of Sciences, Doctor of Medical Sciences, Professor, Director.
111123, Moscow, Novogireevskaya str. 3a
Russian FederationReferences
- Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010; 2(8): 1804-20. https://doi.org/10.3390/v2081803.
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020; 27(3): 325-8. https://doi.org/10.1016/j.chom.2020.02.001.
- Udugama B., Kadhiresan P, Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020; 14(4): 3822-35. https://doi.org/10.1021/acsnano.0c02624.
- Drosten C., Gottig S., Schilling S., Asper M., Panning M., Schmitz H., et al. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, Dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 2002; 40(7): 2323-30. https://doi.org/10.1128/jcm.40.7.2323-2330.2002.
- Mackay I.M. Real-time PCR in virology. Nucleic Acids Research. 2002; 30(6): 1292-305. https://doi.org/10.1093/nar/30.6.1292.
- Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020; 26(7): 771-83. https://doi.org/10.1261/rna.076232.120.
- Shen M., Zhou Y., Ye J., Abdullah Al-Maskri A.A., Kang Y., Zeng S., et al. Recent advances and perspectives of nucleic acid detection for coronavirus. J. Pharm. Anal. 2020; 10(1): 97-101. https://doi.org/10.1016/j.jpha.2020.02.010.
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V, Singh J., et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020; 38(7): 870-4. https://doi.org/10.1038/s41587-020-0513-4.
- Schmid-Burgk J.L., Schmithausen R.M., Li D., Hollstein R., Ben-Shmuel A., Israeli O., et al. LAMP-Seq: population-scale COVID-19 diagnostics using combinatorial barcoding. bioRxiv. 2020.04.06.025635. https://doi.org/10.1101/2020.04.06.025635.
- Hueston L., Kok J., Guibone A., McDonald D., Hone G., Goodwin J., et al. The antibody response to SARS-CoV-2 infection. Open Forum Infect. Dis. 2020; 7(9): ofaa387. https://doi.org/10.1093/ofid/ofaa387.
- Kiselev D., Matsvay A., Abramov I., Dedkov V., Shipulin G., Khafizov K. Current trends in diagnostics of viral infections of unknown etiology. Viruses. 2020; 12(2): 211. https://doi.org/10.3390/v12020211.
- Blanco L., Bemad A., Lazaro J.M., Martin G., Garmendia C., Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem. 1989; 264(15): 8935-40.
- Compton J. Nucleic acid sequence-based amplification. Nature. 1991; 350(6313): 91-2. https://doi.org/10.1038/350091a0.
- Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006; 4(7): e204. https://doi.org/10.1371/journal.pbio.0040204.
- Vincent M., Xu Y., Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004; 5(8): 795-800. https://doi. org/10.1038/sj.embor.7400200.
- Notomi T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Research. 2000; 28(12): E63. https://doi.org/10.1093/nar/28.12.e63.
- Thompson D., Lei Y. Mini review: Recent progress in RT-LAMP enabled COVID-19 detection. Sens. Actuators Rep. 2020; 2(1): 100017. https://doi.org/10.1016/j.snr.2020.100017.
- Francois P., Tangomo M., Hibbs J., Bonetti E.J., Boehme C.C., Notomi T., et al. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med Microbiol. 2011; 62(1): 41-8. https://doi.org/10.1111j.1574-695X.2011.00785.x.
- Rodel J., Egerer R., Suleyman A., Sommer-Schmid B., Baier M., Henke A., et al. Use of the variplex™ SARS-CoV-2 rT-LAMP as a rapid molecular assay to complement RT-PCR for COVID-19 diagnosis. J. Clin. Virol. 2020; 132: 104616. https://doi.org/10.1016/).jcv.2020.104616.
- El-Tholoth M., Branavan M., Naveenathayalan A., Balachandran W. Recombinase polymerase amplification-nucleic acid lateral flow immunoassays for Newcastle disease virus and infectious bronchitis virus detection. Mol. Biol. Rep. 2019; 46: 6391-7. https://doi.org/10.1007/s11033-019-05085-y.
- Lamb L.E., Bartolone S.N., Ward E., Chancellor M.B. Rapid detection of novel coronavirus/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS One. 2020; 15(6): e0234682. https://doi.org/10.1371/journal.pone.0234682.
- Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell Probes. 2002; 16(3): 223-9. https://doi.org/10.1006/mcpr.2002.0415.
- Parida M., Sannarangaiah S., Dash P.K., Rao P.V.L., Morita K. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008; 18(6): 407-21. https://doi.org/10.1002/rmv.593.
- Sahoo P.R., Sethy K., Mohapatra S., Panda D. Loop mediated isothermal amplification: An innovative gene amplification technique for animal diseases. Vet. World. 2016; 9(5): 465-9. https://doi.org/10.14202/vetworld.2016.465-469.
- Mahony J., Chong S., Bulir D., Ruyter A., Mwawasi K., Waltho D. Multiplex loop-mediated isothermal amplification (M-LAMP) assay for the detection of influenza A/H1, A/H3 and influenza B can provide a specimen-to-result diagnosis in 40 min with single genome copy sensitivity. J. Clin. Virol. 2013; 58(1): 127-31. https://doi.org/10.1016/jjcv.2013.06.006.
- Foo P.C., Chan Y.Y., Mohamed M., Wong W.K., Nurul Najian A.B., Lim B.H. Development of a thermostabilised triplex LAMP assay with dry-reagent four target lateral flow dipstick for detection of Entamoeba histolytica and non-pathogenic Entamoeba spp. Anal. Chim. Acta. 2017; 966: 71-80. https://doi.org/10.1016j.aca.2017.02.019.
- Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Com-mun. 2001; 289(1): 150-4. https://doi.org/10.1006/bbrc.2001.5921.
- Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., et al. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin. Chem. 2020; 66(7): 975-7. https://doi.org/10.1093/clinchem/hvaa102.
- Karthik K., Rathore R., Thomas P., Arun T.R., Viswas K.N., Dhama K., et al. New closed tube loop mediated isothermal amplification assay for prevention of product cross-contamination. MethodsX. 2014; 1: 137-43. https://doi.org/10.1016/j.mex.2014.08.009.
- Tanner N.A., Zhang Y., Evans T.C. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques. 2015; 58(2): 59-68. https://doi.org/10.2144/000114253.
- Wastling S.L., Picozzi K., Kakembo A.S.L., Welburn S.C. LAMP for human African trypanosomiasis: a comparative study of detection formats. PLoS Negl. Trop. Dis. 2010; 4(11): e865. https://doi.org/10.1371/journal.pntd.0000865.
- Iwamoto T., Sonobe T., Hayashi K. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J. Clin. Microbiol. 2003; 41(6): 2616-22. https://doi.org/10.1128/jcm.41.6.2616-2622.2003.
- Tomita N., Mori Y., Kanda H., Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008; 3(5): 877-82. https://doi.org/10.1038/nprot.2008.57.
- Gandelman O.A., Church V.L., Moore C.A., Kiddle G., Carne C.A., Parmar S., et al. Novel bioluminescent quantitative detection of nucleic acid amplification in real-time. PLoS One. 2010; 5(11): e14155. https://doi.org/10.1371/journal.pone.0014155.
- Roy S., Wei S.X., Ying J.L.Z., Safavieh M., Ahmed M.U. A novel, sensitive and label-free loop-mediated isothermal amplification detection method for nucleic acids using luminophore dyes. Biosens. Bioelectron. 2016; 86: 346-52. https://doi.org/10.1016j.bi-os.2016.06.065.
- Chou P.H., Lin Y.C., Teng P.H., Chen C.L., Lee P.Y. Real-time target-specific detection of loop-mediated isothermal amplification for white spot syndrome virus using fluorescence energy transfer-based probes. J. Virol. Methods. 2011; 173(1): 67-74. https://doi.org/10.1016/jjviromet.2011.01.009.
- Becherer L., Borst N., Bakheit M., Frischmann S., Zengerle R., von Stetten F. Loop-mediated isothermal amplification (LAMP) -review and classification of methods for sequence-specific detection. Anal. Methods. 2020; 12(6): 717-46. https://doi.org/10.1039/C9AY02246E.
- Nyan D.C., Swinson K.L. A novel multiplex isothermal amplification method for rapid detection and identification of viruses. Sci. Rep. 2015; 5: 17925. https://doi.org/10.1038/srep17925.
- Liu W., Huang S., Liu N., Dong D., Yang Z., Tang Y., et al. Establishment of an accurate and fast detection method using molecular beacons in loop-mediated isothermal amplification assay. Sci. Rep. 2017; 7: 40125. https://doi.org/10.1038/srep40125.
- Wang L., Yang C.J., Medley C.D., Benner S.A., Tan W. Locked nucleic acid molecular beacons. J. Am. Chem. Soc. 2005; 127(45): 15664-5. https://doi.org/10.1021/ja052498g.
- Tani H., Teramura T., Adachi K., Tsuneda S., Kurata S., Nakamura K., et al. Technique for quantitative detection of specific DNA sequences using alternately binding quenching probe competitive assay combined with loop-mediated isothermal amplification. Anal. Chem. 2007; 79(15): 5608-13. https://doi.org/10.1021/ac070041e.
- Nazarenko I., Pires R., Lowe B., Obaidy M., Rashtchian A. Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes. Nucleic Acids Res. 2002; 30(9): 2089-195. https://doi.org/10.1093/nar/30.9.2089.
- Gadkar V.J., Goldfarb D.M., Gantt S., Tilley P.A.G. Real-time detection and monitoring of loop mediated amplification (LAMP) reaction using self-quenching and de-quenching fluorogenic probes. Sci. Rep. 2018; 8(1): 5548. https://doi.org/10.1038/s41598-018-23930-1.
- Ayukawa Y., Hanyuda S., Fujita N., Komatsu K., Arie T. Novel loop-mediated isothermal amplification (LAMP) assay with a universal QProbe can detect SNPs determining races in plant pathogenic fungi. Sci. Rep. 2017; 7(1): 4253. https://doi.org/10.1038/s41598-017-04084-y.
- Zerilli F., Bonanno C., Shehi E., Amicarelli G., Adlerstein D., Makrigiorgos G.M. Methylation-specific loop-mediated isothermal amplification for detecting hypermethylated DNA in simplex and multiplex formats. Clin. Chem. 2010; 56(8): 1287-96. https://doi.org/10.1373/clinchem.2010.143545.
- Kouguchi Y., Fujiwara T., Teramoto M., Kuramoto M. Homogenous, real-time duplex loop-mediated isothermal amplification using a single fluorophore-labeled primer and an intercalator dye: Its application to the simultaneous detection of Shiga toxin genes 1 and 2 in Shiga toxigenic Escherichia coli isolates. Mol. Cell Probes. 2010; 24(4): 190-5. https://doi.org/10.1016/j.mcp.2010.03.001.
- Tanner N.A., Zhang Y., Evans T.C. Jr. Simultaneous multiple target detection in real-time loop-mediated isothermal amplification. Biotechniques. 2012; 53(2): 81-9. https://doi.org/10.2144/0000113902.
- Kubota R., Jenkins D.M. Real-time duplex applications of Loop-mediated AMPlification (LAMP) by assimilating probes. Int. J. Mol. Sci. 2015; 16(3): 4786-99. https://doi.org/10.3390/ijms16034786.
- Jiang Y.S., Bhadra S., Li B., Wu Y.R., Milligan J.N., Ellington A.D. Robust strand exchange reactions for the sequence-specific, real-time detection of nucleic acid amplicons. Anal. Chem. 2015; 87: 3314-20. https://doi.org/10.1021/ac504387c.
- Kumar Y., Bansal S., Jaiswal P. Loop-mediated isothermal amplification (LAMP): A rapid and sensitive tool for quality assessment of meat products. Compr. Rev. Food Sci. Food Saf 2017; 16(6): 1359-78. https://doi.org/10.1111/1541-4337.12309.
- Kidd M., Richter A., Best A., Mirza J., Percival B., Mayhew M., et al. S-variant SARS-CoV-2 is associated with significantly higher viral loads in samples tested by ThermoFisher TaqPath RT-QPCR. medRxiv. 2020; 2020.12.24.20248834. https://doi.org/10.1101/202 0.12.24.20248834.
- Huang W.E., Lim B., Hsu C.C., Xiong D., Wu W., Yu Y, et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020; 13(4): 950-61. https://doi.org/10.1111/1751-7915.13586.
- Park G.S., Ku K., Baek S.H., Kim S.J., Kim S.I., Kim B.T., et al. Development of reverse transcription loop-mediated isothermal amplification assays targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Mol. Diagn. 2020; 22(6): 729-35. https://doi.org/10.1016/jjmoldx.2020.03.006.
- Sun F., Ganguli A., Nguyen J., Brisbin R., Shanmugam K., Hirsch-berg D.L., et al. Smartphone-based multiplex 30-minute nucleic acid test of live virus from nasal swab extract. Lab. Chip. 2020; 20(9): 1621-7. https://doi.org/10.1039/d0lc00304b.
- Jiang M., Pan W., Arasthfer A., Fang W., Ling L., Fang H., et al. Development and validation of a rapid, single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. Front. Cell. Infect. Microbiol. 2020; 10: 331. https://doi.org/10.3389/fcimb.2020.00331.
- Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.P., Kirrmaier D., et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020; 12(556): eabc7075. https://doi.org/10.1126/sci-translmed.abc7075.
- Tian F., Liu C., Deng J., Han Z., Zhang L., Chen Q., et al. A fully automated centrifugal microfluidic system for sample-to-answer viral nucleic acid testing. Sci. China Chem. 2020; 1-9. https://doi.org/10.1007/s11426-020-9800-6.
- Ganguli A., Mostafa A., Berger J., Aydin M.Y., Sun F., de Ramirez S.A.S., et al. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020; 117(37): 22727-35. https://doi.org/10.1073/pnas.2014739117.
- Rohaim M.A., Clayton E., Sahin I., Vilela J., Khalifa M.E., Al-Natour M.Q., et al. Artificial intelligence-assisted loop mediated isothermal amplification (AI-LAMP) for rapid detection of SARS-CoV-2. Viruses. 2020; 12(9): 972. https://doi.org/10.3390/v12090972.
- Xiong H., Ye X., Li Y., Wang L., Zhang J., Fang X., et al. Rapid differential diagnosis of seven human respiratory coronaviruses based on centrifugal microfluidic nucleic acid assay. Anal. Chem. 2020; 92(21): 14297-302. https://doi.org/10.1021/acs.analchem.0c03364.
- Mautner L., Baillie C.K., Herold H.M., Volkwein W., Guertler P., Eberle U., et al. Rapid point-of-care detection of SARS-CoV-2 using reverse transcription loop-mediated isothermal amplification (RT-LAMP). Virol. J. 2020; 17(1): 160. https://doi.org/10.1186/s12985-020-01435-6.
- Mohon A.N., Oberding L., Hundt J., van Marle G., Pabbaraju K., Berenger B.M., et al. Optimization and clinical validation of dual-target RT-LAMP for SARS-CoV-2. J. Virol. Methods. 2020; 286: 113972. https://doi.org/10.1016/jjviromet.2020.113972.
- Schermer B., Fabretti F., Damagnez M., Di Cristanziano V, Heger E., Arjune S., et al. Rapid SARS-CoV-2 testing in primary material based on a novel multiplex RT-LAMP assay. PLoS One. 2020; 15(11): e0238612. https://doi.org/10.1371/journal.pone.0238612.
- Varlamov D.A., Blagodatskikh K.A., Smirnova E.V., Kramarov V.M., Ignatov K.B. Combinations of PCR and isothermal amplification techniques are suitable for fast and sensitive detection of SARS-CoV-2 viral RNA. Front. Bioeng. Biotechnol. 2020; 8: 604793. https://doi.org/10.3389/fbioe.2020.604793.
- Lei Y. Kitchen range oven enabled one-tube RT-LAMP for RNA detection at home - A potential solution for large-scale screening of COVID-19. engrXiv. 2020. Preprint. https://doi.org/10.31224/osf.io/ed85s.
Supplementary files