The features of Tat protein of human immunodeficiency virus type 1 (Retroviridae: Lentivirus: Lentivirus humimdef1) non-A6 variants, characteristic for the Russian Federation
- Authors: Kuznetsova A.I.1, Antonova A.A.1, Lebedev A.V.1, Ozhmegova E.N.1, Shlykova A.V.2, Lapovok I.A.2, Galzitskaya O.V.1,3,4
-
Affiliations:
- D.I. Ivanovsky Institute of Virology of National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya
- Central Research Institute of Epidemiology
- Institute of Protein Research RAS
- Institute of Theoretical and Experimental Biophysics RAS
- Issue: Vol 69, No 6 (2024)
- Pages: 524-534
- Section: ORIGINAL RESEARCHES
- URL: https://journal-vniispk.ru/0507-4088/article/view/277914
- DOI: https://doi.org/10.36233/0507-4088-272
- EDN: https://elibrary.ru/xdvhfq
- ID: 277914
Cite item
Abstract
Introduction. Tat protein is a trans-activator of HIV-1 genome transcription, with additional functions including the ability to induce the chronic inflammatory process. Natural amino acid polymorphisms in Tat may affect its functional properties and the course of HIV infection.
The aim of this work is to analyze the marks of Tat consensus sequences in non-A6 HIV-1 variants characteristic of the Russian Federation, as well as study natural polymorphisms in Tat CRF63_02A6 and subtype B variants circulating in Russia.
Materials and methods. The whole-genome nucleotide sequences of HIV-1 CRF63_02A6, CRF03_A6B, as well as subtype B and CRF02_AG circulating in Russia were used. The reference group was formed based on the sequences of subtype B variants circulating in different countries. Preferentially, the sequences were downloaded from the international database Los Alamos.
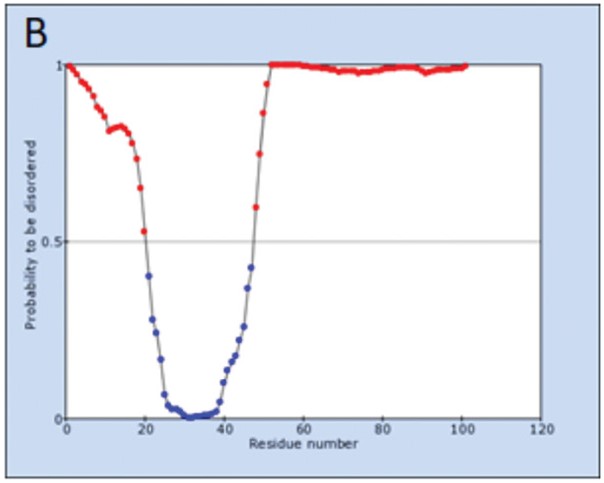
Results. CRF63_02A6 consensus sequence contained the highest number of amino acid substitutions, 31, and had no helix at positions 30‒33 in the secondary structure; however, this did not change its predicted tertiary structure. CRF03_A6B consensus sequence contained a stop codon at position 87. The polymorphisms in subtype B variants circulating in our country and in CRF63_02A6 variants were identified.
Conclusion. Consensus sequences of Tat protein in non-A6 variants typical for the Russian Federation were obtained and their features were determined. R78G, located in the functionally significant motif, and C31S, the functionally significant substitution, were significantly more frequent in subtype B variants circulating in Russia and in CRF63_02A6 variants than in the reference group, respectively. A limitation of this study is the small sample of sequences.
Full Text
##article.viewOnOriginalSite##About the authors
Anna I. Kuznetsova
D.I. Ivanovsky Institute of Virology of National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya
Author for correspondence.
Email: a-myznikova@list.ru
ORCID iD: 0000-0001-5299-3081
head of laboratory of T-lymphotropic viruses, PhD, leading researcher
Russian Federation, 123098, MoscowAnastasiia A. Antonova
D.I. Ivanovsky Institute of Virology of National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya
Email: aantonova1792@gmail.com
ORCID iD: 0000-0002-9180-9846
PhD, Researcher, Laboratory of T-lymphotropic viruses
Russian Federation, 123098, MoscowAleksey V. Lebedev
D.I. Ivanovsky Institute of Virology of National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya
Email: lebedevalesha236@gmail.com
ORCID iD: 0000-0001-6787-9345
PhD, Researcher, Laboratory of T-lymphotropic viruses
Russian Federation, 123098, MoscowEkaterina N. Ozhmegova
D.I. Ivanovsky Institute of Virology of National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya
Email: belokopytova.01@mail.ru
ORCID iD: 0000-0002-3110-0843
PhD, Researcher, Laboratory of T-lymphotropic viruses
Russian Federation, 123098, MoscowAnastasia V. Shlykova
Central Research Institute of Epidemiology
Email: murzakova_a.v@mail.ru
ORCID iD: 0000-0002-1390-8021
Researcher
Russian Federation, Moscow, 111123Ilya A. Lapovok
Central Research Institute of Epidemiology
Email: i_lapovok@mail.ru
ORCID iD: 0000-0002-6328-1415
PhD, Senior researcher
Russian Federation, Moscow, 111123Oxana V. Galzitskaya
D.I. Ivanovsky Institute of Virology of National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya; Institute of Protein Research RAS; Institute of Theoretical and Experimental Biophysics RAS
Email: ogalzit@vega.protres.ru
ORCID iD: 0000-0002-3962-1520
Doctor of Physical and Mathematical Sciences, Head of the Bioinformatics Laboratory, Chief Researcher
Russian Federation, 123098, Moscow; 142290, Moscow Region, Pushchino; 142290, Moscow Region, PushchinoReferences
- German Advisory Committee Blood (Arbeitskreis Blut), Subgroup «Assessment of Pathogens Transmissible by Blood». Human Immunodeficiency Virus (HIV). Transfus. Med. Hemother. 2016; 43(3): 203–22. https://doi.org/10.1159/000445852
- Bbosa N., Kaleebu P., Ssemwanga D. HIV subtype diversity worldwide. Curr. Opin. HIV AIDS. 2019; 14(3): 153–60. https://doi.org/10.1097/COH.0000000000000534
- Kuznetsova A.I. The role of HIV-1 polymorphism in the pathogenesis of the disease. VICh-infektsiya i immunosupressii. 2023; 15(3): 26–37. https://doi.org/10.22328/2077-9828-2023-15-3-26-37 https://elibrary.ru/cwjjai (in Russian)
- Antonova A.A., Kuznetsova A.I., Ozhmegova E.N., Lebedev A.V., Kazennova E.V., Kim K.V., et al. Genetic diversity of HIV-1 at the current stage of the epidemic in the Russian federation: an increase in the prevalence of recombinant forms. VICh-infektsiya i immunosupressii. 2023; 15(3): 61–72. https://doi.org/10.22328/2077-9828-2023-15-3-61-72 https://elibrary.ru/tpwttn (in Russian)
- Kuznetsova A.I., Gromov K.B., Kireev D.E., Shlykova A.V., Lopatukhin A.E., Kazennova E.V., et al. Analysis of tat protein characteristics in human immunodeficiency virus type 1 sub-subtype A6 (Retroviridae: Orthoretrovirinae: lentivirus: human immunodeficiency Virus-1). Voprosy virusologii. 2021; 66(6): 452–63. https://doi.org/10.36233/0507-4088-83 https://elibrary.ru/cmzgyc (in Russian)
- Li L., Dahiya S., Kortagere S., Aiamkitsumrit B., Cunningham D., Pirrone V., et al. Impact of Tat genetic variation on HIV-1 disease. Adv. Virol. 2012; 2012: 123605. https://doi.org/10.1155/2012/123605
- Kuznetsova A., Kim K., Tumanov A., Munchak I., Antonova A., Lebedev A., et al. Features of Tat protein in HIV-1 sub-subtype A6 variants circulating in the Moscow Region, Russia. Viruses. 2023; 15(11): 2212. https://doi.org/10.3390/v15112212
- Ajasin D., Eugenin E.A. HIV-1 Tat: Role in bystander toxicity. Front. Cell. Infect. Microbiol. 2020: 10: 61. https://doi.org/10.3389/fcimb.2020.00061
- Kamori D., Ueno T. HIV-1 Tat and viral latency: what we can learn from naturally occurring sequence variations. Front. Microbiol. 2017; 8: 80. https://doi.org/10.3389/fmicb.2017.00080
- Spector C., Mele A.R., Wigdahl B., Nonnemacher M.R. Genetic variation and function of the HIV-1 Tat protein. Med. Microbiol. Immunol. 2019; 208(2): 131–69. https://doi.org/10.1007/s00430- 019-00583-z
- Ranga U., Shankarappa R., Siddappa N.B., Ramakrishna L., Nagendran R., Mahalingam M., et al. Tat protein of human immunodeficiency virus type 1 subtype C strains is a defective chemokine. J. Virol. 2004; 78(5): 2586–90. https://doi.org/10.1128/jvi.78.5.2586-2590.2004
- Ruiz A.P., Ajasin D.O., Ramasamy S., DesMarais V., Eugenin E.A., Prasad V.R. A naturally occurring polymorphism in the HIV-1 Tat basic domain inhibits uptake by bystander cells and leads to reduced neuroinflammation. Sci. Rep. 2019; 9(1): 3308. https://doi.org/10.1038/s41598-019-39531-5
- Jin H., Li D., Lin M.H., Li L., Harrich D. Tat-based therapies as an adjuvant for an HIV-1 functional cure. Viruses. 2020; 12(4): 415. https://doi.org/10.3390/v12040415
- Asamitsu K., Fujinaga K., Okamoto T. HIV Tat/P-TEFb interaction: a potential target for novel anti-HIV therapies. Molecules. 2018; 23(4): 933. https://doi.org/10.3390/molecules23040933
- Lobanov M.Y., Sokolovskiy I.V., Galzitskaya O.V. IsUnstruct: prediction of the residue status to be ordered or disordered in the protein chain by a method based on the Ising model. J. Biomol. Struct. Dyn. 2013; 31(10): 1034–43. https://doi.org/10.1080/07391102.2012.718529
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021; 596(7873): 583–9. https://doi.org/10.1038/s41586-021-03819-2
- Shafer R.W., Rhee S.Y., Pillay D., Miller V., Sandstrom P., Schapiro J.M., et al. HIV-1 protease and reverse transcriptase mutations for drug resistance surveillance. AIDS. 2007; 21(2): 215–23. https://doi.org/10.1097/QAD.0b013e328011e691
- Berezov T.T., Korovkin B.F. Biological Chemistry [Biologicheskaya khimiya]. Moscow: Meditsina; 1998. (in Russian)
- Lobanov M.Y., Pereyaslavets L.B., Likhachev I.V., Matkarimov B.T., Galzitskaya O.V. Is there an advantageous arrangement of aromatic residues in proteins? Statistical analysis of aromatic interactions in globular proteins. Comput. Struct. Biotechnol. J. 2021; 19: 5960–8. https://doi.org/10.1016/j.csbj.2021.10.036
- Tee K.K., Thomson M.M., Hemelaar J. Editorial: HIV-1 genetic diversity, volume II. Front. Microbiol. 2022; 13: 1007037. https://doi.org/10.3389/fmicb.2022.1007037
- Bobkova M.R. Genetic diversity of human immunodeficiency viruses and antiretroviral therapy. Terapevticheskii arkhiv. 2016; 88(11): 103–11. https://doi.org/10.17116/terarkh20168811103-111 https://elibrary.ru/xeaxsb (in Russian)
- Cilento M.E., Kirby K.A., Sarafianos S.G. Avoiding drug resistance in HIV reverse transcriptase. Chem. Rev. 2021; 121(6): 3271–96. https://doi.org/10.1021/acs.chemrev.0c00967
- Gotora P.T., Brown K., Martin D.R., van der Sluis R., Cloete R., Williams M.E. Impact of subtype C-specific amino acid variants on HIV-1 Tat-TAR interaction: insights from molecular modelling and dynamics. Virol. J. 2024; 21(1): 144. https://doi.org/10.1186/s12985-024-02419-6
- Mishra M., Vetrivel S., Siddappa N.B., Ranga U., Seth P. Clade-specific differences in neurotoxicity of human immunodeficiency virus-1 B and C Tat of human neurons: Significance of dicysteine C30C31 motif. Ann. Neurol. 2008; 63(3): 366–76. https://doi.org/10.1002/ana.21292
Supplementary files