Разработка, изучение и сравнение моделей перекрестного иммунитета к вирусу гриппа с применением статистических методов и машинного обучения
- Авторы: Асатрян М.Н.1, Шмыр И.С.1, Тимофеев Б.И.1, Щербинин Д.Н.1, Агасарян В.Г.1, Тимофеева Т.А.1, Ершов И.Ф.1, Герасимук Э.Р.1,2, Ноздрачева А.В.1, Семененко Т.А.1, Логунов Д.Ю.1, Гинцбург А.Л.1
-
Учреждения:
- ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи»
- ФГБОУ ВО «Университет «Дубна»
- Выпуск: Том 69, № 4 (2024)
- Страницы: 349-362
- Раздел: ОРИГИНАЛЬНЫЕ ИССЛЕДОВАНИЯ
- URL: https://journal-vniispk.ru/0507-4088/article/view/265972
- DOI: https://doi.org/10.36233/0507-4088-250
- EDN: https://elibrary.ru/phejeu
- ID: 265972
Цитировать
Аннотация
Введение. Всемирная организация здравоохранения в качестве одного из важнейших критериев оценки успешно проводимой вакцинации и способности предотвращать заболевание у населения рассматривает значения титров антител в реакции торможения гемагглютинации. Математическое моделирование перекрестного иммунитета позволяет оперативно выявлять новые антигенные варианты, что имеет первостепенное значение для эпидемиологического надзора и здоровья человека.
Материалы и методы. В настоящей работе применены статистические методы и техники машинного обучения от простого к сложному ‒ регрессионная логистическая модель, метод случайного леса и градиентный бустинг. В расчетах, параллельно дистанции Хемминга, также использовали матрицы AАindex. Вычисления проводили с разными типами и величинами порогов антигенного ускользания, на четырех наборах данных (временны́х периодах). Результаты сравнивали по принятым метрикам бинарной классификации.
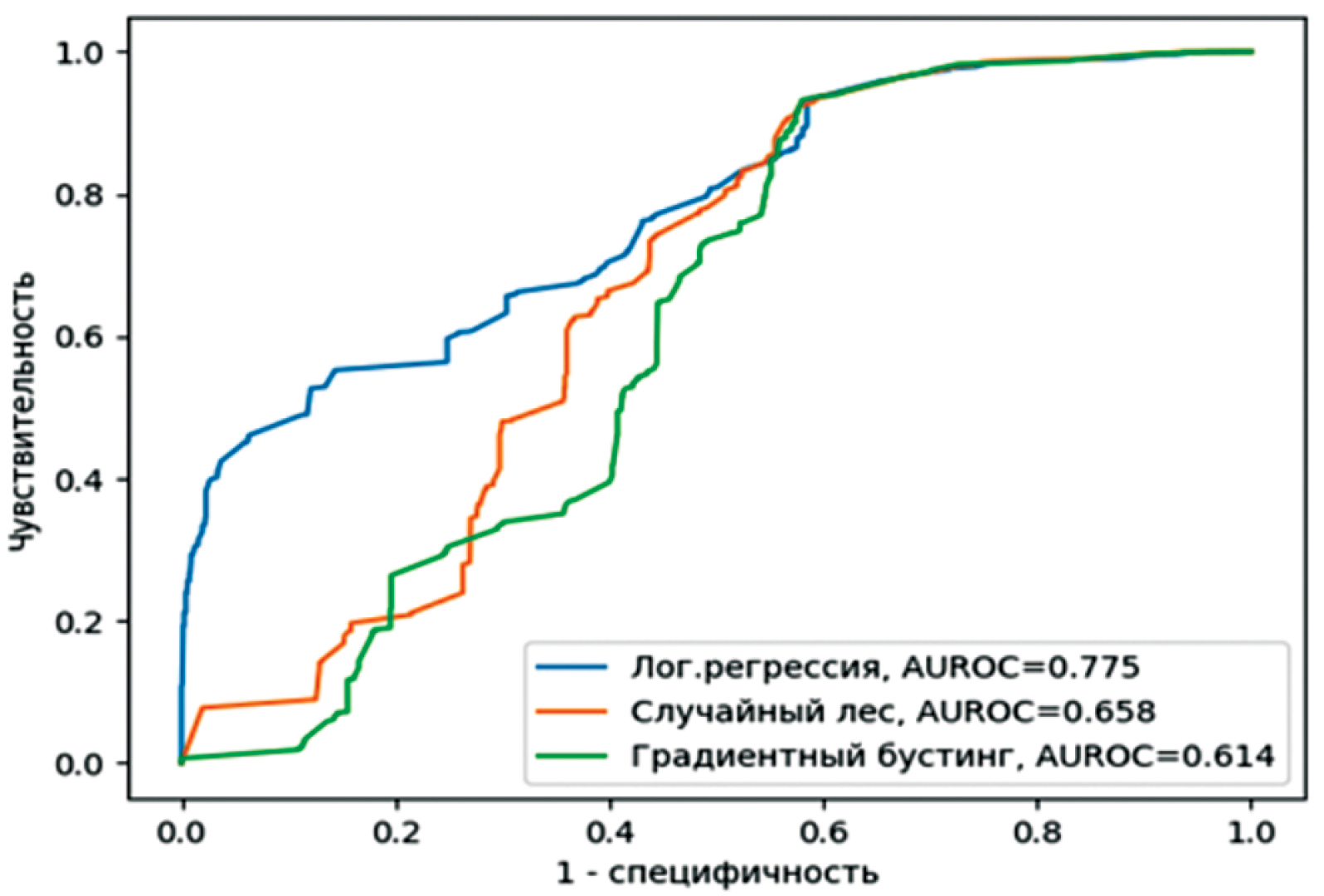
Результаты. Показана существенная дифференциация в зависимости от применяемых наборов данных. Лучшие результаты продемонстрировали все три модели на прогнозный осенний сезон 2022 г., предварительно обученные на февральском сезоне этого же года (AUROC 0,934; 0,958; 0,956 соответственно). Наименьшие результаты были получены на весь прогнозный 2023 г., настроенные на данных двух сезонов 2022 г. (AUCROC 0,614; 0,658; 0,775 соответственно). При этом зависимость результатов от применяемых типов порогов и их величин оказалась незначительной. Дополнительное применение матриц ААindex не улучшило существенно результаты моделей, но в то же время не внесло значимых ухудшений.
Заключение. Более сложные модели показывают лучший результат. При разработке моделей перекрестного иммунитета, для убедительного утверждения об их прогностической устойчивости важно проводить тестирование на разных наборах данных.
Ключевые слова
Полный текст
Открыть статью на сайте журналаОб авторах
Марина Норайровна Асатрян
ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи»
Автор, ответственный за переписку.
Email: masatryan@gamaleya.org
ORCID iD: 0000-0001-6273-8615
канд. мед. наук, старший научный сотрудник группы эпидемиологической кибернетики отдела эпидемиологии
Россия, 123098, г. МоскваИлья Сергеевич Шмыр
ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи»
Email: shmyris@gamaleya.org
ORCID iD: 0000-0002-8514-5174
научный сотрудник группы эпидемиологической кибернетики отдела эпидемиологии
Россия, 123098, г. МоскваБорис Игоревич Тимофеев
ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи»
Email: timofeevbi@gamaleya.org
ORCID iD: 0000-0001-7425-0457
‒ канд. физ.-мат. наук, старший научный сотрудник лаборатории физиологии вирусов Института вирусологии им. Д.И. Ивановского
Россия, 123098, г. МоскваДмитрий Николаевич Щербинин
ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи»
Email: shcherbinindn@gamaleya.org
ORCID iD: 0000-0002-8518-1669
канд. биол. наук, старший научный сотрудник отдела генетики и молекулярной биологии бактерий
Россия, 123098, г. МоскваВаагн Гагикович Агасарян
ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи»
Email: agasaryanvg@gamaleya.org
ORCID iD: 0009-0009-3824-7061
научный сотрудник группы эпидемиологической кибернетики отдела эпидемиологии
Россия, 123098, г. МоскваТатьяна Анатольевна Тимофеева
ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи»
Email: timofeeva.tatyana@gamaleya.org
ORCID iD: 0000-0002-8991-8525
канд. биол. наук, заведующая лабораторией физиологии вирусов Института вирусологии им. Д.И. Ивановского
Россия, 123098, г. МоскваИван Феликсович Ершов
ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи»
Email: ershovif@gamaleya.org
ORCID iD: 0000-0002-3333-5347
научный сотрудник группы эпидемиологической кибернетики отдела эпидемиологии
Россия, 123098, г. МоскваЭлита Русиндапутри Герасимук
ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи»; ФГБОУ ВО «Университет «Дубна»
Email: ealita@mail.ru
ORCID iD: 0000-0002-7364-163X
канд. мед. наук, доцент
Россия, 123098, г. Москва; 141982, г. ДубнаАнна Валерьевна Ноздрачева
ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи»
Email: nozdrachevaav@gamaleya.org
ORCID iD: 0000-0002-8521-1741
канд. мед. наук, заведующая лабораторией неспецифической профилактики инфекционных заболеваний отдела эпидемиологии
Россия, 123098, г. МоскваТатьяна Анатольевна Семененко
ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи»
Email: semenenko@gamaleya.org
ORCID iD: 0000-0002-6686-9011
д-р мед. наук, профессор, академик РАЕН, главный научный сотрудник отдела эпидемиологии
Россия, 123098, г. МоскваДенис Юрьевич Логунов
ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи»
Email: logunov@gamaleya.org
ORCID iD: 0000-0003-4035-6581
д-р биол. наук, академик РАН, заместитель директора по научной работе
Россия, 123098, г. МоскваАлександр Леонидович Гинцбург
ФГБУ «Национальный исследовательский центр эпидемиологии и микробиологии имени почетного академика Н.Ф. Гамалеи»
Email: gintsburg@gamaleya.org
ORCID iD: 0000-0003-1769-5059
д-р биол. наук, профессор, академик РАН, директор
Россия, 123098, г. МоскваСписок литературы
- Walker P.J., Siddell S.G., Lefkowitz E.J., Mushegian A.R., Adriaenssens E.M., Alfenas-Zerbini P., et al. Recent changes to viruses taxonomy ratified by the International Committee on Taxonomy of Viruses. Arch. Virol. 2022; 167(11): 2429–40. https://doi.org/10.1007/s00705-022-05516-5
- Chen J., Li K., Rong H., Bilal K., Yang N., Li K. A disease diagnosis and treatment recommendation system based on big data mining and cloud computing. Inf. Sci. 2018; 435: 124–49. https://doi.org/10.1016/j.ins.2018.01.001
- Qiu J., Qiu T., Yang Y., Wu D., Cao Z. Incorporating structure context of HA protein to improve antigenicity calculation for influenza virus A/H3N2. Sci. Rep. 2016; 6: 31156. https://doi.org/10.1038/srep31156
- Асатрян М.Н., Агасарян В.Г, Щербинин Д.Н., Тимофеев Б.И., Ершов И.Ф., Шмыр И.С. и др. Influenza IDE. Патент РФ № 2020617965; 2020.
- Асатрян М.Н., Тимофеев Б.И., Шмыр И.С., Хачатрян К.Р., Щербинин Д.Н., Тимофеева Т.А. и др. Математическая модель для оценки уровня перекрёстного иммунитета между штаммами вируса гриппа подтипа H3N2. Вопросы вирусологии. 2023; 68(3): 252–64. https://doi.org/10.36233/0507-4088-179 https://elibrary.ru/rexvea
- Nakai K., Kidera A., Kanehisa M. Cluster analysis of amino acid indices for prediction of protein structure and function. Protein Eng. 1988; 2(2): 93–100. https://doi.org/10.1093/protein/2.2.93
- Virology Research Services. The Hemagglutination Inhibition Assay; 2023. Available at: https://virologyresearchservices.com/2023/04/07/understanding-the-hai-assay/
- Spackman E., Sitaras I. Hemagglutination Inhibition Assay. In: Animal Influenza Virus. 2020; 11–28. Available at: https://link.springer.com/protocol/10.1007/978-1-0716-0346-8_2
- Kaufmann L., Syedbasha M., Vogt D., Hollenstein Y., Hartmann J., Linnik J.E., et al. An optimized Hemagglutination Inhibition (HI) assay to quantify influenza-specific antibody titers. J. Vis Exp. 2017; (130): 55833. https://doi.org/10.3791/55833
- Burnet F.M., Lush D. The action of certain surface active agents on viruses. Aust. J. Exp. Biol. Med. Sci. 1940; 18(2): 141–50.
- Bedford T., Suchard M.A., Lemey P., Dudas G., Gregory V., Hay A.J., et al. Integrating influenza antigenic dynamics with molecular evolution. Elife. 2014; 3: e01914. https://doi.org/10.7554/eLife.01914
- Anderson C.S., McCall P.R., Stern H.A., Yang H., Topham D.J. Antigenic cartography of H1N1 influenza viruses using sequence-based antigenic distance calculation. BMC Bioinformatics. 2018; 19(1): 51. https://doi.org/10.1186/s12859-018-2042-4
- Lee M.S., Chen J.S. Predicting antigenic variants of influenza A/H3N2 viruses. Emerg. Infect. Dis. 2004; 10(8): 1385–90. https://doi.org/10.3201/eid1008.040107
- МУ 3.1.3490–17. Изучение популяционного иммунитета к гриппу у населения Российской Федерации: Методические указания; 2017.
- Lin X., Lin F., Liang T., Ducatez M.F., Zanin M., Wong S.S. Antibody responsiveness to influenza: what drives it? Viruses. 2021; 13(7): 1400. https://doi.org/10.3390/v13071400
- Lees W.D., Moss D.S., Shepherd A.J. A computational analysis of the antigenic properties of haemagglutinin in influenza A H3N2. Bioinformatics. 2010; 26(11): 1403–8. https://doi.org/10.1093/bioinformatics/btq160
- Zhou X., Yin R., Kwoh C.K., Zheng J. A context-free encoding scheme of protein sequences for predicting antigenicity of diverse influenza A viruses. BMC Genomics. 2018; 19(Suppl. 10): 936. https://doi.org/10.1186/s12864-018-5282-9
- Peng Y., Wang D., Wang J., Li K., Tan Z., Shu Y., et al. A universal computational model for predicting antigenic variants of influenza A virus based on conserved antigenic structures. Sci. Rep. 2017; 7: 42051. https://doi.org/10.1038/srep42051
- Huang J.W., Yang J.M. Changed epitopes drive the antigenic drift for influenza A (H3N2) viruses. BMC Bioinformatics. 2011; 12(Suppl. 1): S31. https://doi.org/10.1186/1471-2105-12-S1-S31
- Tolles J., Meurer W.J. Logistic regression: relating patient characteristics to outcomes. JAMA. 2016; 316(5): 533–4. https://doi.org/10.1001/jama.2016.7653
- Hastie T., Tibshirani R., Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Springer; 2009.
- Zacour М., Ward В.J., Brewer A., Tang P., Boivin G., Li Y. Standardization of hemagglutination inhibition assay for influenza serology allows for high reproducibility between laboratories. Clin. Vaccine Immunol. 2016; 23(3): 236–42. https://doi.org/10.1128/CVI.00613-15
- Кильбурн Э.Д., ред. Вирусы гриппа и грипп. Пер. с англ. М.: Медицина; 1978.
- Yao Y., Li X., Liao B., Huang L., He P., Wang F., et al. Predicting influenza antigenicity from Hemagglutintin sequence data based on a joint random forest method. Sci. Rep. 2017; 7(1): 1545. https://doi.org/10.1038/s41598-017-01699-z
- Lee E.K., Tian H., Nakaya H.I. Antigenicity prediction and vaccine recommendation of human influenza virus A (H3N2) using convolutional neural networks. Hum. Vaccin. Immunother. 2020; 16(11): 2690–708. https://doi.org/10.1080/21645515.2020.1734397
- Shah S.A.W., Palomar D.P., Barr I., Poon L.L.M., Quadeer A.A., McKay M.R. Seasonal antigenic prediction of influenza A H3N2 using machine learning. Nat. Commun. 2024; 15(1): 3833. https://doi.org/10.21203/rs.3.rs-2924528/v1
- Wang P., Zhu W., Liao B., Cai L., Peng L., Yang J. Predicting influenza antigenicity by matrix completion with antigen and antiserum similarity. Front. Microbiol. 2018; 9: 2500. https://doi.org/10.3389/fmicb.2018.02500
- Huang L., Li X., Guo P., Yao Y., Liao B., Zhang W., et al. Matrix completion with side information and its applications in predicting the antigenicity of influenza viruses. Bioinformatics. 2017; 33(20): 3195–201. https://doi.org/ 10.1093/bioinformatics/btx390
- Liao Y.C., Lee M.S., Ko C.Y., Chao A.H. Bioinformatics models for predicting antigenic variants of influenza A/H3N2 virus. Bioinformatics. 2008; 24(4): 505–12. https://doi.org/10.1093/bioinformatics/btm638
- Yang J., Zhang T., Wan X.F. Sequence-based antigenic change prediction by a sparse learning method incorporating co-evolutionary information. PLoS One. 2014; 9(9): e106660. https://doi.org/10.1371/journal.pone.0106660
- Adabor E.S. A statistical analysis of antigenic similarity among influenza A (H3N2) viruses. Heliyon. 2021; 7(11): e08384. https://doi.org/10.1016/j.heliyon.2021.e08384
Дополнительные файлы