Molecular methods for diagnosing novel coronavirus infection: comparison of loop-mediated isothermal amplification and polymerase chain reaction
- Authors: Akimkin V.G.1, Petrov V.V.1, Krasovitov K.V.1, Borisova N.I.1, Kotov I.A.1, Rodionova E.N.1, Cherkashina A.S.1, Kondrasheva L.Y.1, Tivanova E.V.1, Khafizov K.F.1
-
Affiliations:
- FSBI «Central Research Institute for Epidemiology» of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
- Issue: Vol 66, No 6 (2021)
- Pages: 417-424
- Section: ORIGINAL RESEARCHES
- URL: https://journal-vniispk.ru/0507-4088/article/view/118199
- DOI: https://doi.org/10.36233/0507-4088-86
- ID: 118199
Cite item
Abstract
Introduction. Currently, the basis for molecular diagnostics of most infections is the use of reverse transcription polymerase chain reaction (RT-PCR). Technologies based on reverse transcription isothermal loop amplification (RT-LAMP) can be used as an alternative to RT-PCR for diagnostic purposes. In this study, we compared the RTLAMP and RT-PCR methods in order to analyze both the advantages and disadvantages of the two approaches.
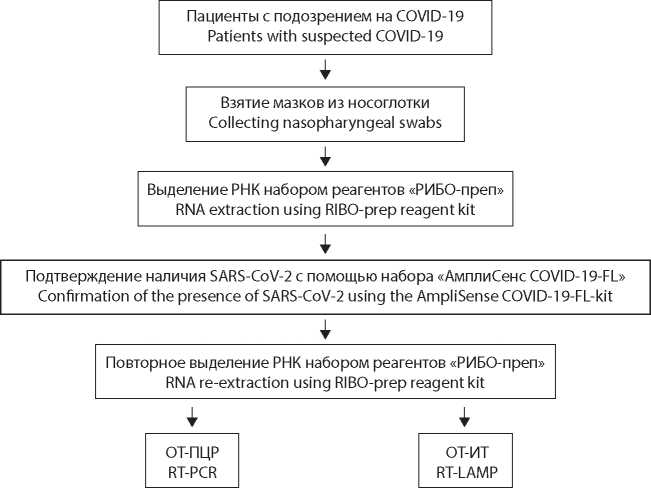
Material and methods. For the study, we used reagent kits based on RT-PCR and RT-LAMP. The biological material obtained by taking swabs from the mucous membrane of the oropharynx and nasopharynx in patients with symptoms of a new coronavirus infection was used.
Results. We tested 381 RNA samples of the SARS-CoV-2 virus (Coronaviridae: Coronavirinae: Betacoronavirus; Sarbecovirus) from various patients. The obtained values of the threshold cycle (Ct) for RT-PCR averaged 20.0 ± 3.7 s (1530 ± 300 s), and for RT-LAMP 12.8 ± 3.7 s (550 ± 160 s). Proceeding from the theoretical assumptions, a linear relationship between values obtained in two kits was proposed as a hypothesis; the correlation coefficient was approximately 0.827. At the same time, for samples with a low viral load (VL), the higher Ct values in RT-LAMP did not always correlated with those obtained in RT-PCR.
Discussion. We noted a significant gain in time for analysis using RT-LAMP compared to RT-PCR, which can be important in the context of testing a large number of samples. Being easy to use and boasting short turnaround time, RT-LAMP-based test systems can be used for mass screening in order to identify persons with medium and high VLs who pose the greatest threat of the spread of SARS-CoV-2, while RT-PCR-based diagnostic methods are also suitable for estimation of VL and its dynamics in patients with COVID-19.
Full Text
##article.viewOnOriginalSite##About the authors
V. G. Akimkin
FSBI «Central Research Institute for Epidemiology» of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
ORCID iD: 0000-0003-4228-9044
Moscow, 111123, Russia
Russian FederationV. V. Petrov
FSBI «Central Research Institute for Epidemiology» of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
ORCID iD: 0000-0002-3503-2366
Moscow, 111123, Russia
Russian FederationK. V. Krasovitov
FSBI «Central Research Institute for Epidemiology» of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
ORCID iD: 0000-0001-7237-1810
Moscow, 111123, Russia
Russian FederationN. I. Borisova
FSBI «Central Research Institute for Epidemiology» of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
ORCID iD: 0000-0002-9672-0648
Moscow, 111123, Russia
Russian FederationI. A. Kotov
FSBI «Central Research Institute for Epidemiology» of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
ORCID iD: 0000-0003-2416-5689
Moscow, 111123, Russia
Russian FederationE. N. Rodionova
FSBI «Central Research Institute for Epidemiology» of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
ORCID iD: 0000-0003-0192-1832
Moscow, 111123, Russia
Russian FederationA. S. Cherkashina
FSBI «Central Research Institute for Epidemiology» of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
ORCID iD: 0000-0002-1888-4903
Moscow, 111123, Russia
Russian FederationL. Yu. Kondrasheva
FSBI «Central Research Institute for Epidemiology» of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
ORCID iD: 0000-0002-0147-4262
Moscow, 111123, Russia
Russian FederationE. V. Tivanova
FSBI «Central Research Institute for Epidemiology» of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
ORCID iD: 0000-0003-1286-2612
Moscow, 111123, Russia
Russian FederationK. F. Khafizov
FSBI «Central Research Institute for Epidemiology» of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Author for correspondence.
Email: kkhafizov@gmail.com
ORCID iD: 0000-0001-5524-0296
Kamil’ F. Khafizov, Ph.D. (Biol.), Head, Scientific Group for the Development of New Diagnostic Methods, Molecular Diagnostics and Epidemiology Department
Moscow, 111123, Russia
Russian FederationReferences
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020; 323(13): 1239–42. https://doi.org/10.1001/jama.2020.2648
- Sharfstein J.M., Becker S.J., Mello M.M. Diagnostic Testing for the Novel Coronavirus. JAMA. 2020; 323(15): 1437–8. https://doi.org/10.1001/jama.2020.3864
- Drosten C., Göttig S., Schilling S., Asper M., Panning M., Schmitz H., et al. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 2002; 40(4): 2323–30. https://doi.org/10.1128/jcm.40.7.2323-2330.2002
- Mackay I.M., Arden K.E., Nitsche A. Real-time PCR in virology. Nucleic Acids Res. 2002; 30(6): 1292–305. https://doi.org/10.1093/nar/30.6.1292
- Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020; 20(5): 453–4. https://doi.org/10.1080/14737159.2020.1757437
- Lu R., Wu X., Wan Z., Li Y., Zuo L., Qin J., et al. Development of a novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Virol. Sin. 2020; 35(3):344–7. https://doi.org/10.1007/s12250-020-00218-1
- Jiang M., Pan W., Arasthfer A., Fang W., Ling L., Fang H., et al. Development and validation of a rapid, single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. Front. Cell Infect. Microbiol. 2020; 10: 331. https://doi.org/10.3389/fcimb.2020.00331
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000; 28(12): E63. https://doi.org/10.1093/nar/28.12.e63
- Хафизов К.Ф., Петров В.В., Красовитов К.В., Золкина М.В., Акимкин В.Г. Экспресс-диагностика новой коронавирусной инфекции с помощью реакции петлевой изотермической амплификации. Вопросы вирусологии. 2021; 66(1): 17–28. https://doi.org/10.36233/0507-4088-42
- Parida M., Posadas G., Inoue S., Hasebe F., Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 2004; 42(1):257–63. https://doi.org/10.1128/jcm.42.1.257-263.2004
- Zhao Y., Chen F., Li Q., Wang L., Fan C. Isothermal amplification of nucleic acids. Chem. Rev. 2015; 115(22): 12491–545. https://doi.org/10.1021/acs.chemrev.5b00428
- Bruno A., de Mora D., Freire-Paspuel B., Rodriguez A.S., Paredes- Espinosa M.B., Olmedo M., et al. Analytical and clinical evaluation of a heat shock SARS-CoV-2 detection method without RNA extraction for N and E genes RT-qPCR. Int. J. Infect. Dis. 2021; 109: 315–20. https://doi.org/10.1016/j.ijid.2021.06.038
- Lalli M.A., Langmade J.S., Chen X., Fronick C.C., Sawyer C.S., Burcea L.C., et al. Rapid and Extraction-Free Detection of SARS-CoV-2 from Saliva by Colorimetric Reverse-Transcription Loop-Mediated Isothermal Amplification. Clin. Chem. 2021; 67(2):415–24. https://doi.org/10.1093/clinchem/hvaa267
- Anastasiou O.E., Holtkamp C., Schäfer M., Schön F., Eis-Hübinger A.M., Krumbholz A. Fast Detection of SARS-CoV-2 RNA Directly from Respiratory Samples Using a Loop-Mediated Isothermal Amplification (LAMP) Test. Viruses. 2021; 13. https://doi.org/10.3390/v13050801
- Thompson D., Lei Y. Mini review: Recent progress in RT-LAMP enabled COVID-19 detection. Sensors and Actuators Reports. 2020; 2: 100017. http://dx.doi.org/10.1016/j.snr.2020.100017
- Borisova N.I., Kotov I.A., Kolesnikov A.A., Kaptelova V.V., Speranskaya A.S., Kondrasheva L.Yu., et al. Monitoring the spread of the SARS-CoV-2 (Coronaviridae: Coronavirinae: Betacoronavirus; Sarbecovirus) variants in the Moscow region using targeted high-throughput sequencing. Voprosy Virusologii. 2021; 66(4):269–78. https://doi.org/10.36233/0507-4088-72
- Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021; 2(1): e13–22. https://doi.org/10.1016/s2666-5247(20)30172-5
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020; 395(10229):1054–62. https://doi.org/10.1016/s0140-6736(20)30566-3
- La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020; 39(6): 1059–61. https://doi.org/10.1007/s10096-020-03913-9
- Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin. Infect. Dis. 2020; 71(10):2663–6. https://doi.org/10.1093/cid/ciaa638
- Mora-Cárdenas E., Marcello A. Switch-on the LAMP to spot Zika. Ann. Transl. Med. 2017; 5(24): 500. https://doi.org/10.21037/atm.2017.10.19
- Augustine R., Hasan A., Das S., Ahmed R., Mori Y., Notomi T., et al. Loop-mediated isothermal amplification (LAMP): A rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology (Basel). 2020; 9(8): 182. https://doi.org/10.3390/biology9080182
- Rabe B.A., Cepko C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl. Acad. Sci. USA. 2020; 117(39):24450–8. https://doi.org/10.1073/pnas.2011221117
- Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., et al. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin. Chem. 2020; 66(7): 975–7. https://doi.org/10.1093/clinchem/hvaa102
Supplementary files